Kaiyi Jiang (姜凯议)
Biological Engineer
Massachusetts Institute of Technology
Biography
I am currently a Ph.D. candidate at Massachusetts Institute of Technology (MIT) Biological Engineering. I am fortunate to be advised by Dr. Jonathan Gootenberg, Dr. Omar Abudayyeh, and Dr. Michael Birnbaum. Previously, I received my B.S. (Summa cum laude) in biomedical engineering from Rice University in 2021. At Rice, I was fortunate to work with Dr. Caleb Bashor and Dr. Gang Bao. I was an intern at Regeneron Pharmaceutical’s therapeutic antibody group in 2018. I am broadly interested in harnessing biological diversity to discover novel reprogrammable systems and using machine learning to engineer these systems for programmable cell control and delivery. Applications include in vivo recording and lineage tracing, gene and cell therapy for autoimmune disease and cancer.
- Gene/Cell Therapy
- Synthetic Biology
- AI/ML for Biology
PhD in Biological Engineering, 2025
Massachusetts Institute of Technology
BS in Biomedical Engineering, 2021
Rice University
Experience
- Developed the first robust mammalian cell RNA sensor named reprogrammable ADAR sensors (RADARS) that senses endogenous RNA transcripts down to 13TPM and release arbitrary payload upon ADAR mediated stop codon editing. I demonstrated this technology for cell state-specific apoptosis, molecular recording (lineage tracing with CRE), in vivo detection of tissue markers with live bioluminescence imaging, and RNA gated synthetic mRNA cytokine therapies for RNA immunotherapy in solid tumor.
- Built a low-N protein engineering ensemble model comprising large protein langue model (ESM) and domain specific expert top layer for rapid evolution of enzymatic function. This model achieved SOTA performance on public DMS datasets. I deployed this model on a novel miniature CRISPR nuclease (PsaCas12f) and rapidly evolved a 10-fold more active enPsaCas12f for in vivo genome editing. Using this model, I evolved a SOTA T7 RNA polymerase that produces mRNA with near zero immunogenicity, BXB1 integrase that are 2-fold more active than wild type, and carbonic anhydrase with 20% increased thermal and PH stability.
- Discovered the first RNA-guided protease in type III-E CRISPR systems (Craspase Cas7-11/Csx29/Csx30). This is the first known abortive infection module in bacterial antiviral defense systems that utilize post-translational protein cleavage upon RNA detection. I reprogrammed the system to function as a RNA sensor system in mammalian cell and demonstrated the potential for mammalian cell RNA diagnostic and therapy.
- Discovered a novel group of eukaryotic RNA-guided DNA endonucleases (Fanzor). This is the first example of RNA-guided DNA cleavage mechanism in eukaryotes, and demonstrated the powerful evolution of RNA-guided nuclease TnpB’s adaptation into the eukaryotic world as they gradually acquired NLS and introns. Bioinformatic mining revealed more than 3,000 novel clusters in the eukaryotic genomes and serve as a rich resource for future nucleases.
Featured Publications
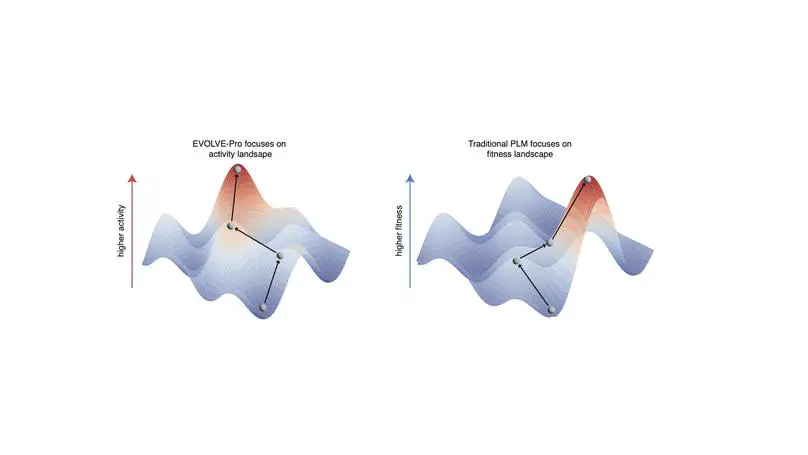
We developed a novel machine learning model to rapidly evolve protein in silico for higher activity. We demonstrated SOTA performance for EVOLVEpro across DMS benchamarks and used it evolve a highly active genome editing toolbox and T7 RNAP that are orders of magnitude higher fiedlity than current used wild-type.
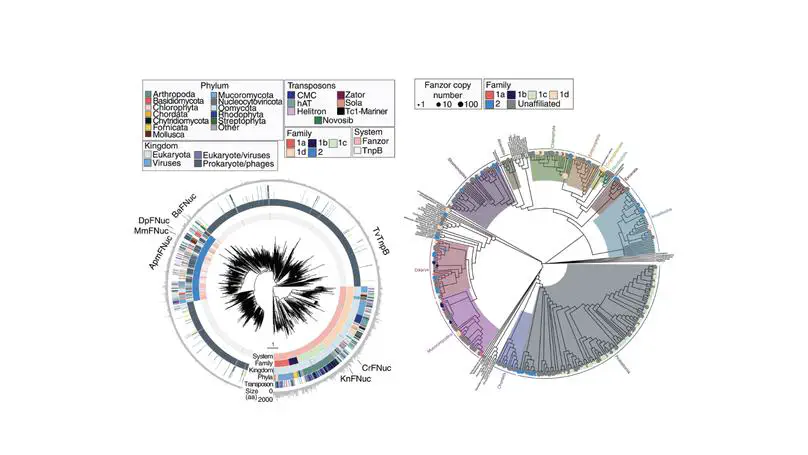
We discovered the first eukaryotic RNA-guided nucleases that are homologs of bacterial TnpB and CRISPR/Cas12 nucleases. This work revealed novel evolutionary insights on RNA-guided systems in eukaryotes. In addition, we performed comprehensive biochemical characeterization on fanzor’s adaption in eukaryotes and engineered it for mammalian cell genome editing.
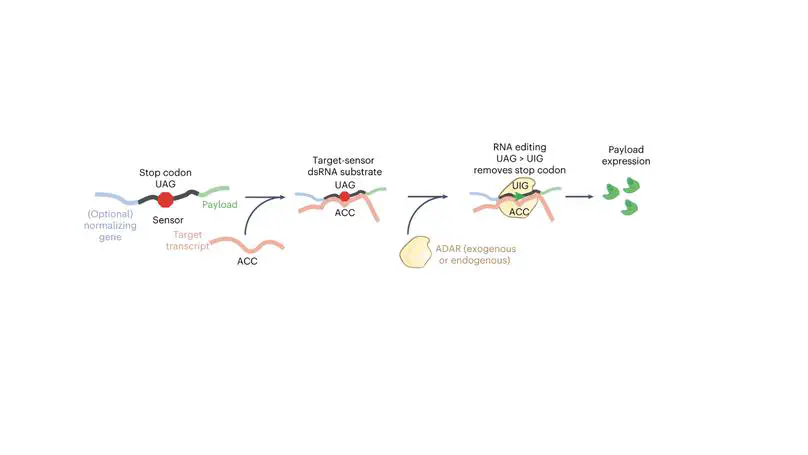
We engineered the first robust mammalian RNA-sensor based on ADAR called RADARS. The sensor can be reprogrammed to track any RNA species inside eukaryotic cells and allow conditional cargo expression based on the presence/expression of target mRNA(s). We demonstrate that the system can be readily integrated into AAV, lentivirus, and synthetic mRNA to selectively turn on an arbitrary protein of interest. We showcase the use of this system in cell specific killing, lineage tracing and in vivo recording for reprogrammable cell control.
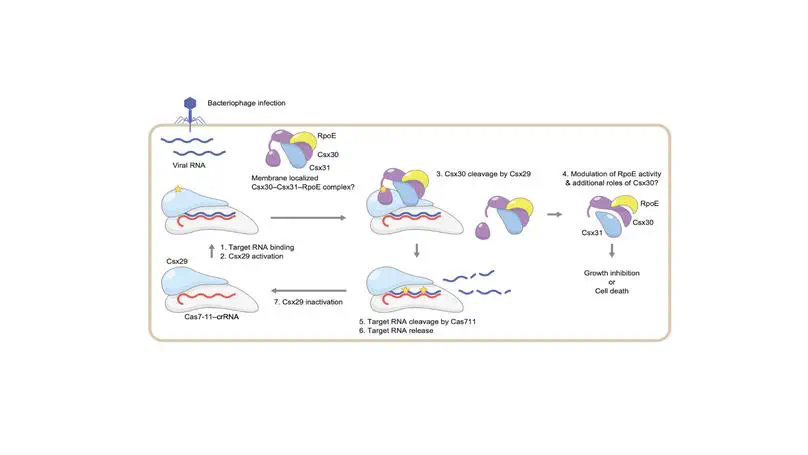
We discovered the first RNA-guided protease systems in prokaryotic antiviral defense systems. We biochemically characterized the Cas7-11/Csx29/Csx30 systems in the context of abortive infection module against phage invasion. We then engineered the system and adapted it as an RNA-sensor system in mammalian cell.